What Happens When Tame Meets Wild? Texas Scientists Look to Ducks for Answers
It’s an ecological and agricultural problem noted in many species. For example, when fish are bred in tanks, they are all fed and the majority survive. But when those same fish are released into the wild, recessive traits can explode — with significant negative consequences for the overall population.
What happens when domesticated and wild animals interbreed is a research focus for Philip Lavretsky, Assistant Professor of Biological Sciences at the University of Texas at El Paso. He uses mallard ducks as a model system because of their unique behavior and evolutionary history. For the past 20 years, the eastern population of mallards has been declining steadily, making for an excellent case study of the adaptive impacts of hybridization.
Lavretsky’s team uses a combination of next-generation sequencing techniques to generate genome-wide markers and to link genetic variation to species or population traits of interest. For a close look at population genetics at the landscape level with hundreds of thousands of samples, the team employs a double digest restriction-site associated DNA sequencing (ddRADseq) methodology on the Illumina HiSeq® systems for partial genome sequencing. To understand which genes may be important in the survival of the species, they use Oxford Nanopore sequencing technology to look at 125 full genome sequences. In addition, the team is using ancient DNA techniques to analyze hundreds of preserved mallards from museum collections to track traits present in today’s population.
“We are looking closely at the genomes for genetic variation that may be important in the adaptive potential of wild mallards, while also uncovering mechanisms that may explain some of the recent declining trends in North American mallards,” Lavretsky told us. “Specifically, if they cannot eat, they cannot survive through the winter and migrate as well as nest properly, and so with enough generations of less fit individuals you expect a population decline.”
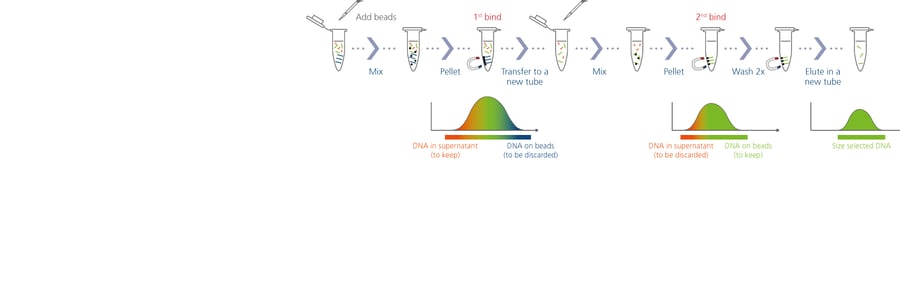
For this work, the team is using the Quantabio sparQ PureMag Beads, a fast and reliable nucleic acid purification system, to optimize the size selection and PCR cleanup steps for both the ddRADseq application on the Illumina sequencers and the PCR cleanup with the Oxford Nanopore protocols. Prior to incorporating this product, Lavretsky’s team had to spend two months analyzing 120 samples. With the new optimized protocols, they are now able to manually run 250 samples in only 16 hours, with a 38% cost savings compared to the previous workflow.
Additionally, the team has transitioned to utilizing AccuStart Long Range SuperMix, a PCR master mix that enables easy, long range amplification of genomic target regions (up to 24 kb), with high accuracy. Employed in Whole Genomic Sequencing with Oxford Nanopore Technology, this AccuStart Long Range SuperMix has been instrumental in uncovering significant structural rearrangements (inversions) within the Wild Type Mallard Duck Genome compared to Game Farm Mallard and Khaki Campbell Mallards. This finding underscores the pivotal role of breed-specific artificial selection that occurs due to genomic divergence as a result of domestication of these animals.
To learn more about the team’s work, check out our customer profile. For more information about using sparQ PureMag Beads for size selection, read this application note.
Recent posts


Subscribe to Our Blog
Read More

Metagenomics Express: New Study Reports Faster Library Prep for 16S rRNA Analysis
App Notes: Versatility for DNA and RNA Cleanup