Double Duty: Our Library Prep Kits Streamline QC for PCR-Free Sequencing
Good news: Quantabio is now offering a discount on our sparQ DNA Library Prep Kit and our sparQ DNA Frag & Library Prep Kit. This promotion will last through the rest of 2024, so you’ll have plenty of time to stock up. Contact your sales representative to find out the highly competitive prices available in your location.
These library prep products are handy for a broad range of uses. Library quality control, for example, is an essential step in next-generation sequencing (NGS) workflows to ensure successful sequencing outcomes. It’s through QC analysis that we check for DNA fragment length, yield, library size distribution, and the presence of impurities such as adapter dimers or other unwanted DNA fragments. A gold standard method for library size distribution QC is the use of automated electrophoresis, where DNA fragment migration serves as visual confirmation of sample quality.
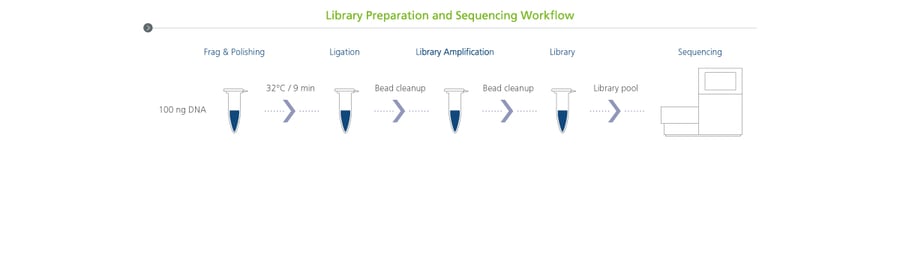
One common library preparation method is to use a PCR-free library protocol, which can be helpful when sequencing genomic regions with biased base composition (high GC or AT content). The elimination of the PCR amplification step from the NGS workflow reduces nucleotide misincorporation and improves genome assembly, but the QC analysis of these libraries presents some challenges. PCR-free libraries with Y-shaped adapters migrate at a slower rate during electrophoresis due to their open and single-stranded ends. As a result, these libraries require additional analysis to generate accurate QC information.
We have developed a method to conduct an accurate QC analysis of PCR-free libraries using the sparQ DNA Frag & Library Prep Kit or the sparQ DNA Library Prep Kit, without incurring additional cost for library preparation. Here’s how it works: we use the HiFi PCR Master Mix and Primer Mix (included with the sparQ DNA library prep kits) to run a short PCR amplification on an aliquot of 1 μl of the PCR-free library before analyzing the PCR product on an electrophoretic system. PCR cycling linearizes the library fragments by generating complementary double-stranded ends from the template libraries. This leads to optimal migration and the QC step reflects the true fragment size of the PCR-free library.
This approach offers a cost-effective and accurate alternative, streamlining the process of generating and verifying PCR-free libraries for NGS analysis. Library size distribution and quality can easily be determined using widely available QC platforms. Then, once the size distribution has been properly characterized, qPCR library quantification analysis can be performed to accurately determine the concentration and yield of the PCR-free libraries as the final QC step before sequencing.
Check out our application note for a detailed protocol and to check out results.
Recent posts


Subscribe to Our Blog
Read More

Now Available: Step-by-Step Video Tutorials for sparQ NGS Products
App Note: Rapid Library Preparation for RNA Virus Sequencing